This topic takes on average 55 minutes to read.
There are a number of interactive features in this resource:
 Biology
Biology
 Chemistry
Chemistry
 PSHE / Citizenship studies
PSHE / Citizenship studies
 Science
Science
Mutations can occur randomly during DNA replication. Examples include addition, deletion and substitution of nucleotide bases.
If the mutation occurs in a gene, it can alter the amino acid sequence coded. Modifying the amino acid sequence of a polypeptide chain can have various consequences including modifying protein structure. Hydrogen, ionic and disulphide bonds hold the tertiary and quaternary structures of proteins together. Amino acid changes can disrupt these bonds- or even create new bonds. This can transform the structure of a protein. Structural modifications of a protein can change the shape of its active site, change its function, or stop it functioning.
If the mutation occurs within a non-coding gene promoter region, it can result in increased or decreased levels of protein expression. These mutations can influence the frequency of transcription by altering the affinity of RNA polymerase for the sequence.
Mutations confer antimicrobial resistance in exactly the ways described above. Microbes can be resistant to multiple antimicrobials, often due to multiple resistance mechanisms. Specific mechanisms are shown in the animations below.
Mutations are identified via DNA sequencing. When DNA sequencing first began, scientists did each step manually. Sanger sequencing was the first automated method for DNA sequencing. Sanger sequencing uses a chain termination technique. The target DNA is copied many times in different length fragments. Fluorescently labelled chain terminating nucleotides mark the end of each fragment. Subsequent fluorescence detection enables the sequence of the DNA to be determined.
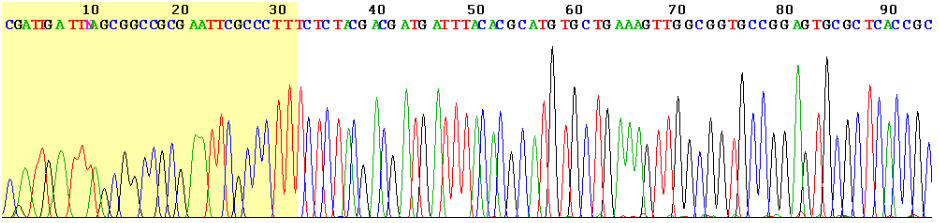
Since the introduction of Sanger sequencing there have been many advancements in sequencing technologies. In 2014, Oxford Nanopore Technologies introduced a new strand sequencing technique. This technique differs from Sanger sequencing as it enables significantly longer fragments of DNA to be sequenced.
Central to the technique are nanopores, which are nano-scale protein holes in membranes. These nanopores have ionic currents passing through them. During sequencing, a single strand of DNA is passed through the nanopore. This changes the ionic current; changes are specific to the nucleotide passing through. These changes are measured and the sequence is subsequently determined.
An example of the nanopore technology is the MinION- which is a pocket sized device costing £900. This device is revolutionary as unlike previous sequencing techniques it can be used everywhere. In 2016, NASA astronaut Kate Rubins used the MinION on the International Space Station- she became the first person to sequence DNA in Space!
Kate Rubins background as a biology researcher enabled her to become a NASA astronaut. For more information on careers in science have a look at our job case studies on the ABPI website.
